CRISPR-Cas9 editing in Plasmodium falciparum
Published:
This is my protocol for using a commercial Cas9 nuclease and custom synthesised guide RNA for CRISPR editing in P. falciparum. I don’t go into how I design the repair template, but to see some examples for HA-tagging and gene disruption, check out my lab book on Benchling.
Credit to Simon Cobbold for first trialling this technique using Integrated DNA Technologies (IDT) reagents. His protocol is available through the IDT website.
Step 1: Designing the guide
Parameters to keep in mind
- I make the homology arms as close as possible to the Cas9 cut site (3 nt upstream from the PAM). I have successfully gone as far as 80 nt away in either direction, however I don’t have any real evidence that closer = better.
- The guide sequence must be absent or mutated in the repair template. In the case of mutation, I’ve found that mutating the PAM, with 4+ mutations in the 20 nt guide-binding sequence are sufficient. (In my preprint, the guide sequence was absent in the final constructs for both the knockouts and HA-tag sequences).
CHOPCHOP and ordering instructions
Follow this link to CHOPCHOP.
- Search for the Target: PlasmoDB ID In: Plasmodium falciparum For knockout
- Click on a “green” guide (high ranked) closest to your desired insertion site.
- Select the Target sequence for the guide, without the PAM (ie, without the NGG - last three nucleotides)
- Go to the IDT guide RNA design site and select Species = other. Paste in the 20 nt guide and click “Check”.
- Check the amount of guide and click “Quick order”. For each guide I buy crRNA, 2 nmol tube.
- Purchase Alt-R® CRISPR-Cas9 tracrRNA, 5 nmol (Cat # 1072532) and Alt-R® S.p. Cas9 Nuclease V3, 100 µg (Cat # 1081058).
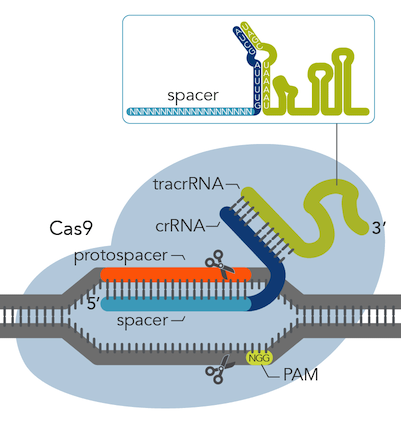
This image from IDT shows how the crRNA and tracrRNA anneal and target the Cas9 to your desired cut site.
Step 2: Transfection
The day before
- To linearise set up an overnight restriction digest with 100 µg plasmid DNA plus 2 or 3 restriction enzymes that will cut the backbone of your repair template plasmid.
- Make sure parasites will be >5% rings the next day
Transfection day
- Run 1 µl of the overnight digest on a gel to make sure it went as expected
- Precipitate the rest of the overnight digest as follows (no gel clean up!):
- add 1/10 volume 3M sodium acetate (pH 5.2)
- Add 2.5 volumes (including sodium acetate volume) of 100% ethanol (mol biol. grade)
- Incubate on ice for >30 mins
- Centrifuge at max speed at 4C for 30 minutes
- Discard supernatant and rinse pellet in 70% ethanol (mol. biol grade)
- Centrifuge at max speed at 4C for 15 minutes
- Open tube in TC hood, remove as much ethanol, and let the plasmid air dry
- Resuspend in 30 µl TE, then add 370 µl cytomix (see recipe below)
- Resuspend tracrRNA and crRNA at 100 µM each in sterile TE.
- Mix 5 µl tracrRNA and 5 µl crRNA in a tube and heat at 95C for 5 min to form gRNA
- Allow to cool to room temp
- Mix 3 ul of gRNA with 2 µl Cas9 Nuclease V3 and leave at room temp for 20 min
- Spin down parasites (need at least 200 µl infected RBCs at >5% rings)
- Add this 5 µl gRNA:Cas9 mixture to the 400 µl cytomix/repair template mixture
- Add 200 µl infected RBCs to the gRNA:Cas9/cytomix/repair
- Transfer to cuvette and electroporate on
- Exponential phase
- 310V
- 950 µF
- infinity resistance
- 2 mm cuvette
- Transfer cuvette contents to a fresh 10 mL dish with 300 µl fresh uninfected RBCs and fresh RPMI.
- Add drug (selection, if applicable) the next day.
- Replace media daily for the first 5 days.
Cytomix (final concentrations)
- 25 mM HEPES/2 mM EGTA, pH 7.6
- 10 mM potassium phosphate pH 7.6
- 120 mM KCl
- 5 mM MgCl2
- 150 mM CaCl2
- Adjust pH to 7.6 with 1M KOH

